Free Access
Table III
Assessment age, number of trees,
number of families, least-squares mean, phenotypic variance, within-subrace additive
variance, narrow-sense heritability ( ) and coefficient
of additive genetic variation (CVa) for
each trait.
) and coefficient
of additive genetic variation (CVa) for
each trait.
| Trait | Assessment | Number | Number | Least-squares | Phenotypic | Additive |
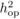
|
C V a | |
|---|---|---|---|---|---|---|---|---|---|
| age (y) | of trees | of families | mean (SE) | variance (SE) | variance (SE) | (SE) | (%) | ||
| Whole-tree volume under bark (m3) | 15 | 246 | 116 | 0.568 (0.016) | 0.049 (0.005) | 0.023 (0.011)* | 0.48 (0.21) | 26.9 | |
| Survival (prop.)† | 15 | 3740 | 381 | 0.528 (0.022) | 1.19 (0.07) | 0.482 (0.068)*** | 0.40 (0.05) | NA | |
| Whole-tree basic density (kg m−3) | 15 | 159 | 102 | 542 (2) | 1034 (127.6) | 947 (359)** | 0.92 (0.29) | 5.7 | |
| Quartersawn board Janka hardness (kN) | 15 | 228 | 115 | 3.77 (0.06) | 0.57 (0.06) | 0.33 (0.144)** | 0.58 (0.23) | 15.2 | |
| Internal checking (prop.)† | – Quartersawn | 15 | 170 | 104 | 0.33 (0.04) | 1.10 (0.17) | 0.25 (0.42)ns | 0.23 (0.35) | NA |
| – Backsawn | 15 | 245 | 116 | 0.18 (0.03) | 1.36 (0.19) | 0.90 (0.47)* | 0.66 (0.25) | NA | |
| DBH15 (cm) | 15 | 1967 | 368 | 25.51 (0.43) | 33.94 (1.13) | 6.08 (1.63)*** | 0.18 (0.05) | 9.7 | |
| Sample basic density (kg m−3) | 15 | 250 | 116 | 513 (2) | 1288 (123.9) | 749 (306)** | 0.58 (0.21) | 5.3 | |
| Sample gross shrinkage (%) | 15 | 250 | 116 | 28.38 (0.40) | 18.97 (1.779) | 6.9 (4.2)* | 0.36 (0.21) | 9.2 | |
| DBH4 (cm) | 4 | 3558 | 381 | 10.95 (0.35) | 8.06 (0.21) | 2.97 (0.37)*** | 0.37 (0.04) | 15.7 | |
| Pilodyn penetration (mm) | 5½ | 1024 | 379 | 13.90 (0.19) | 2.43 (0.12) | 1.15 (0.26)*** | 0.47 (0.10) | 7.7 | |


