| Issue |
Ann. For. Sci.
Volume 67, Number 7, October-November 2010
|
|
|---|---|---|
| Article Number | 702 | |
| Number of page(s) | 9 | |
| DOI | https://doi.org/10.1051/forest/2010027 | |
| Published online | 05 October 2010 | |
Original article
Genetic consequences of harvest in a mature second-growth stand of black walnut (Juglans nigra L.)
1
Department of Forestry and Natural Resources, Hardwood Tree
Improvement and Regeneration Center (HTIRC), Purdue University,
715 West State St, Pfendler Hall,
West Lafayette, IN
47907-2061,
USA
2
Laboratory of Genetics, University of Wisconsin,
Madison,
WI, 53706-1580, USA
3
Department of Forestry and Natural Resources, Purdue
University, West
Lafayette, IN
47907-2033,
USA
4
USDA Forest Service, Hardwood Tree Improvement and Regeneration
Center (HTIRC), Department of Forestry and Natural Resources, Purdue
University, West
Lafayette, IN
47907-2061,
USA
* Corresponding author: rod@hartmannsplantcompany.com
Received:
6
August
2009
Accepted:
15
January
2010
• This study investigated the short-term changes in neutral genetic variation that would occur if a mature, second-growth, black walnut (Juglans nigra L.) stand was harvested following either a diameter-limited or value-limited cutting regime.
• This research tested whether or not the neutral genetic variation within a natural stand of black walnut was disproportionately distributed within the larger or more valuable cohort of trees. To test this hypothesis a complete census of 278 trees from a black walnut stand in Indiana was genotyped using 12 highly polymorphic microsatellites.
• Two types of simulated harvests were performed: (1) diameter-limit cuts, where every tree above a given diameter was removed; and (2) selective harvests based on value, where subsets of the most valuable trees were removed. Allelic diversity of the remaining population after each simulated harvest was compared to a corresponding distribution of outcomes from 10000 random harvests of equal intensity.
• None of the simulated harvests resulted in a reduction of allelic diversity significantly greater than that expected under random harvest, indicating that the allelic diversity of the population was evenly distributed across the entire population.
• These results indicated that typical harvest scenarios do not per se lead to loss of allelic diversity among trees for this species.
Key words: forest genetics / allelic richness / genetic diversity / diameter-limit / high grading
© INRA, EDP Sciences, 2010
1. Introduction
Over the past 200 y, loss of habitat, over-harvesting, and insufficient regeneration have dramatically altered populations of some the most valuable tree species in the central hardwood region (Finkeldey and Ziehe, 2004; Glaubitz et al., 2004; McGuire et al., 1999; Nyland, 2005). In the Midwestern United States, intense land-clearing for agriculture and timber harvesting during the 1800 s and the early part of the 20th century left a highly reduced and fragmented forest landscape. For example, in pre-settlement Indiana, forests were estimated to cover 85% of the state, but by the turn of the twentieth century forest cover had dropped to 13% (Bratkovich et al., 2004). Since this nadir, the trend has reversed to some extent, with about 20% of the state now forested in mature, even-aged stands (Tormoehlen et al., 2000). The small proportion that remains of the once-vast Midwestern forest has been subjected to repeated rounds of selective harvest.
Today, selective harvest remains widespread in this region. The motive is short-term profit: high-grading (value-based harvests) and diameter-limit cuts (the removal of the largest size classes of trees) generally provide the greatest short-term financial gain to land owners, with minimal disturbance to their forests. These selective harvest practices can degrade the forest and decrease the long term economic returns of the residual stand by preventing the release of understory trees, the removal of culls, and natural regeneration (Clatterbuck, 2006; Kenefic et al., 2005; Nyland, 2005).
Many stand and species level studies have focused on the ecological and silvicultural impacts of forest management practices (Angers et al., 2005; Kelty et al., 2003; Kenefic et al., 2005; Nyland, 2005; Tritton and Sendak, 1995). Comparisons of before-and-after treatment, and of treated sites adjacent to undisturbed “control” sites, have been the main approaches used to investigate these questions (Buchert et al., 1997; Glaubitz et al., 2004; Lee et al., 2002). Studies investigating the genetic consequences of forest management have concentrated on economically important gymnosperms (Buchert et al., 1997; Neale, 1985; Rajora, 1999) or tropical tree species (Degen et al., 2006; Lee et al., 2002; Murawski et al., 1994; Obayashi et al., 2002), but few have addressed temperate or fine hardwoods (Glaubitz et al., 2003; Ziehe and Hattemer, 2002). Black walnut was the focal species of this study because it is one of the most valuable hardwood tree species in the eastern and central hardwood regions of the United States, and because it has been the center of a hardwood tree improvement program at Purdue University since the mid-1960s (Victory et al., 2004). Black walnut is shade intolerant, and generally occurs as widely dispersed individuals or in small, spatially distinct groves (Rink et al., 1994). The effect of management on the genetics of black walnut has been the subject of speculation and controversy in the Central Hardwood Region for some time (Beineke, 1974).
Studies investigating the genetic consequences of different silvicultural methods have generally relied on neutral genetic markers due to the difficulty of measuring genetic change in multi-locus adaptive traits. Though neutral genetic markers may not reflect evolutionary changes to loci that undergo selection, they have been found to be useful tools in measuring changes to forest genetic structure due to anthropogenic effects (Finkeldey and Ziehe, 2004). Some of the earliest studies of the genetic consequences of harvest practices utilized allozymes to evaluate genetic change after simulated and actual harvests. In an allozyme study involving a simulated diameter-limit harvest of beech (Fagus sylvatica) trees sampled from a mature German forest, the simulated removal of trees > 55 cm, roughly a 12% census reduction (n ≈ 900), yielded no particular trends and only minor differences in genetic diversity between the removed and remnant trees (Ziehe and Hattemer, 2002). In a Douglas-fir (Pseudotsuga menziesii) shelterwood regeneration study using ten allozyme markers, an approximately 85% reduction of the parent cohorts led to very little loss of allelic diversity in two post-harvested regenerating populations; a result partly attributed to high levels of effective pollen migration and large effective population size (Neale, 1985). An examination of the consequences of silvicultural treatment on the genetics of dogwood (Cornus florida), a shade-tolerant, insect-pollinated, temperate deciduous hardwood tree, showed that “generic concern that silvicultural treatment per se may negatively affect future genetic diversity or increase reproductive isolation is contraindicated” (Sork et al., 2005). Degen et al. (2006) investigated the impact of selective logging on genetic composition and demographic structure of four tropical tree species, and found that “the differences between the genetic parameters in the control scenarios without logging and the logging scenarios were surprisingly small”, mainly because of overlapping generations and effective pollen dispersal, which guaranteed homogeneity of genetic structures of different age classes.
In contrast, Buchert et al. (1997) found that logging reduced the genetic diversity and number of alleles in a white pine (Pinus strobus) stand, and rare alleles were especially reduced. Hawley et al. (2005), in a study of eastern hemlock (Tsuga canadensis), found that diameter-limit thinning resulted in residual populations that had dramatically higher genetic diversity and altered genetic structure. Differences between treatments and controls in the frequencies of rare and uncommon alleles were substantial and likely the underlying cause of genetic diversity differences. They postulated an association between poor phenotypes and rare alleles, a conclusion also supported by Nijensohn et al. (2005), who also observed that removal of the oldest trees in a stand significantly reduced observed heterozygosity. Local spatial genetic structure was found to be essentially random in a second-growth white pine forest, probably as a consequence of previous logging; patches of low-level structure were only observed in an old-growth forest (Marquardt and Epperson, 2004).
Although numerous types of molecular markers have been utilized to answer population genetic questions (Parker et al., 1998), nuclear microsatellites have become the marker of choice over the past decade due to their ease of use and their discerning power. Nuclear microsatellites have been used for the identification of bottlenecks (Aldrich and Hamrick, 1998), to study the genetic consequences of fragmentation (Aldrich et al., 2005; Collevatti et al., 2001), to study the genetic impacts of management practices (Glaubitz et al., 2003), to evaluate genetic structure (Victory et al., 2006), to determine parentage and paternity (Jones and Ardren, 2003; Streiff et al., 1999), to estimate mating success (Dow and Ashley, 1998) and rates of hybridization (Aldrich et al., 2003), and for genetic fingerprinting (Rajora and Rahman, 2003).
The purpose of this study was to examine the short-term changes in neutral genetic variation that would result if a mature, second-growth black walnut stand was harvested following either a diameter-limit or a value-limited cutting regime. Our null hypothesis was that the removal of the largest, most valuable trees (i.e. the founders of the study population) would not significantly alter the stand level allelic richness relative to random removal of the same number of trees. To test the null hypothesis, population level allelic richness (Ar) and other genetic parameters were assessed for a complete census of the stand based on 12 highly polymorphic nuclear microsatellites, and we compared the loss of allelic diversity that would occur under (simulated) selective harvests with the expected loss if a harvest of the same number of stems was carried out at random with respect to value or diameter. Allelic richness has been shown to be the most sensitive measure of genetic erosion resulting from habitat loss, fragmentation, or silvicultural practices (Boshier, 2000; Buchert et al., 1997; Rajora et al., 2000).
2. Materials and methods
2.1. Study site location and sampling techniques
The study was carried out in a stand known as Long Hollow, located in the Hoosier National Forest, Crawford County, Indiana (38° 14′ N latitude/ − 86° 29′ W longitude) (Fig. 1A). The site is a well-drained bottomland that runs mostly in an east - west direction; it is flanked on either side by relatively steep slopes rising about 100 m. The forest is a mature second-growth, even-aged, mixed hardwood stand. Historically, the site was privately owned and used for pasture and timber production. The state of Indiana acquired the land in the mid-1920s and donated it to the USDA Forest Service in 1936 (Dale Weigel, personal communication). Every black walnut (n = 278) within the approximately 97 ha study area was measured for diameter at breast height (dbh) and mapped by geopositioning using a Garmin GPS unit (Garmin, Olathe, KS)1. The raw GPS data were analyzed using arcgis version 9.0 to create a detailed map of the entire black walnut population (Fig. 1B). In addition, mature leaves were sampled from every tree in the population by launching a line up into the canopy by slingshot and pulling down a small branch. The leaf samples were bagged, labeled, and quickly placed on ice. Once in the lab, the leaves were processed for DNA extraction and stored in a − 80 °C freezer, or freeze-dried for future DNA isolation.
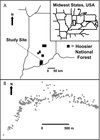 |
Figure 1 (A) Map showing the Long Hollow study area in the Hoosier National Forest HNF, southern Indiana, USA, (inset shows larger view of the area). (B) Location of all 278 black walnut trees sampled at the Long Hollow research site. |
2.2. DNA isolation
Samples were prepared for DNA isolation by grinding about 100 mg of leaf tissue in a 2-ml microcentrifuge tube containing a 1/4-inch cylindrical ceramic bead (BIO 101-Savant, Carlsbad, CA)1 and 1.0 ml of CTAB extraction buffer (Lefort and Douglas, 1999) modified with 2X PVP, 2X CTAB, and 2.0% ß-mercaptoethanol. Samples were homogenized in an FP 120 Fastprep machine (BIO 101-Savant) by grinding for 40 s and then cooling them on ice for about one minute. This cycle was repeated a total of three times. After grinding, the microcentrifuge tubes were placed into a 64 °C water bath for at least 30 min and periodically shaken by hand. Next, samples were centrifuged for 5 min at 12500 × g using a tabletop centrifuge. DNA was isolated from 500 μL of the supernatant using an NA-2000 automated nucleic acid extractor (Autogen, Framingham, MA) employing a modification of Autogen’s NA-2000 Plant DNA, V 1.01 DNA isolation protocol. In this modification, potassium acetate (Autogen reagent AG00317) was added first, followed by SDS/N-lauroyl sarcosine (Autogen reagent AG00212), and then chloroform (Autogen reagent AG00316). All the remaining reagents and protocols were the same as designated in the Autogen protocol. The extracted DNA was quantified using a FL 600 microplate fluorescence reader (Bio-Tek, Winooski, VT) and a Hoechst dye 33258 assay solution. The quantified DNA was diluted with 10 mM Tris-HCl (pH 8.0) and 1.0 mM EDTA (pH 8.0) buffer to a working stock concentration of 10 ng/μL prior to PCR amplification.
2.3. Microsatellite loci and PCR amplification
Twelve nuclear microsatellites developed in our lab were used to genotype the black walnut trees (Tab. I). Detailed information on the twelve loci can found elsewhere (Victory et al., 2006). PCR amplification was performed using Sigma ReadyMix Taq (PCR reaction mix with MgCl2). Each reaction contained 10 ng of DNA template, 5.0 μL ReadyMix Taq PCR reaction mix with MgCl2, 0.4 μL of 20 pmoles/μL working primer stock, and 4 μL of nanopure, sterile water for a total volume of 10 μL. The PCR-amplification protocol was 30 cycles of 92 °C for 30 s, individual annealing temperature for 30 s (see Tab. I), and 72 °C for 30 s, followed by a final incubation at 60 °C for 45 min, run on either a PTC-100TM or a PTC-200TM Peltier Thermal Cycler (MJ Research, INC., San Francisco, CA).
Properties of the 12 microsatellite markers employed in this study. Genbank accession* numbers are in parentheses under the locus name. The number in parentheses under the allele size range is the number of alleles found for that locus in the Long Hollow black walnut population. The FIS values for the black walnut population is based on a total census of the Long Hollow stand.
2.4. Microsatellite analysis and whole population estimates
In preparation for gel electrophoresis, 1.0 μL of the PCR product, 0.5 μL of CXR 400 bp Ladder Standard (Promega, Fitchburg Center, WI) and 1.5 μL of blue dextran loading solution (Promega) were combined, denatured for 2 min at 95 °C, snap cooled with ice, and loaded onto CAL96 paper combs (The Gel Company, San Francisco, CA). Four loci were run together in a single gel lane (multiplexed) by using three differently-labeled fluorescent tags and by ensuring that loci with the same colored tag had alleles with widely differing electrophoretic mobility. For quality control, allelic size standards for each locus were run in three locations, two lanes each, equidistant across the gel to standardize allele calling among gels. Electrophoresis was in 5% polyacrylamide Long Ranger denaturing gels (BMA, Rockland, ME) at 3000 V, 60 mA, 200 W, 51 °C for 3 h using an ABI 377 XL automated DNA sequencer (Perkin-Elmer) with 36-cm plates and 0.2-mm spacers. The software programs genescan v 3.1 and genotyper v 2.5 were used to aid in the assignment of genotypes. The software program gda v 1.1 (Lewis and Zaykin, 2001) was used to calculate the observed number of alleles per locus (allelic richness), the observed and expected heterozygosities, and the fixation index (FIS) for individual loci and across the population based upon the complete census of 278 trees. No exact test was run on the individual loci to make sure they were in Hardy-Weinberg equilibrium (HWE) because we made a complete census of the population. The Long Hollow population estimate of FIS was recalculated by bootstrapping across all loci 10000 times to determine if it significantly deviated from zero. The cumulative number of alleles across all loci were counted and graphically displayed relative to sample size of cohorts created based on size (dbh) and value, starting from the largest diameter cohort (or most valuable trees) and proceeding to the smallest diameter cohorts (or least valuable trees) to determine whether levels of neutral genetic diversity within the population were concentrated in the oldest or most valuable individuals. All of the cohorts based on diameter were subdivided by 5 cm increments, except for the smallest diameter size class (the seedling cohort) which by convention is defined as < 3 cm in diameter. A total of 15 size cohorts was generated by the incremental division based on diameter for the Long Hollow population. All of the cohorts based on value were subdivided by $100 (US dollars) increments. A total of 24 value cohorts was generated by subdividing the Long Hollow population based on the $100 increments.
2.5. Tree grading and value estimation
To simulate a value-based harvest, three experienced professional foresters, skilled in grading forest stands in Indiana, graded every black walnut tree in Long Hollow with a dbh ≥ 30 cm (202 trees) on March 21, 2003. Each forester independently estimated the number of harvestable logs from each tree and assigned each log a grade (not shown). Gross volume in board feet for each tree was calculated using the tree’s dbh, the estimated number of harvestable logs, and an international 1/4-inch rule volume chart. A dollar value was assigned to each tree based on its estimated volume and grade using the 2003 Forest Product Report for the state of Indiana (http://www.ces.purdue.edu/extmedia/FNR/FNR-177-W.pdf). Based on the independent results from each forester, an average value was calculated for each of the 202 black walnut trees surveyed and the values summed to determine an overall value for the population.
2.6. Simulated harvest and data analysis
To examine the effects of different harvest practices on the allelic diversity of a mature, second growth black walnut stand, a series of simulated harvests were performed, including (1) diameter-limit cuts, with limits of 30, 40, 50 or 60 cm (Fig. 2), and (2) selective harvests based on value, where the 10, 15, 25 or 33 percent most valuable trees were removed (Fig. 3). After each simulated harvest, allelic diversity was recalculated based on the genotypes of the residual black walnut trees. The statistical significance of the resulting reduction in allelic richness (one-tailed test) was then determined for the null hypothesis of no relationship between diameter class and allelic richness (or between value class and allelic richness). For each selective harvest, a corresponding null distribution of residual allelic richness (see Fig. 4 for an example) was obtained by performing 10000 random harvests of equal intensity (each with an equal number of “harvested” trees, but chosen at random with respect to diameter or value).
 |
Figure 2 Diameter breast height class distribution of 278 black walnut trees in Long Hollow. Four simulated diameter-limit harvests (30, 40, 50, 60 cm) were performed, removing all individuals to the right of the arrows. Right axis indicates cumulative number of alleles in the population. |
 |
Figure 3 Distribution of the estimated value of the Long Hollow black walnuts. Four simulated value-based harvests were performed by removing the most-valuable 10, 15, 25, and 33% of all the trees from the population. Right axis indicates cumulative number of alleles found in the Long Hollow population. |
 |
Figure 4 An example null distribution of allelic richness (Ar) for the simulated diameter-limit harvest of > 50 cm. The distribution was generated by performing 10000 random harvests of equal intensity each with an equal number of “harvested” trees chosen at random with respect to diameter. After each simulated harvest, allelic diversity was recalculated based on the genotypes of the residual black walnuts, and the observed value for the > 50 cm harvest was then compared to this null distribution to obtain its statistical significance (one-tail test). All other simulated diameter-limit and value-based harvests generated similarly-shaped distributions and comparable P values, so only this example distribution is shown. The shadowed part of the histogram represents the amount of allelic richness (Ar) of the residual trees after the > 50 cm harvest was conducted. |
Weir and Cockerham’s (1984) estimator of population differentiation θ, as well as Nei’s 1972 (D) genetic distance were calculated using gda v 1.1 to compare the levels of genetic divergence and relatedness between the residual trees after each simulated harvest and the pre-harvested population. Significance testing in gda was conducted by bootstrapping 1000 times over all loci. Fisher’s exact test was used to test if the proportion of private alleles differed among cohorts.
3. Results
A high level of genetic diversity was found within the population of 278 black walnuts at Long Hollow based on this set of 12 microsatellites. Across all 12 loci, the overall observed heterozygosity was 0.792, just slightly less than the expected level 0.808, reflecting a slight but non-significant heterozygote deficiency (FIS = 0.020). Analysis of the distribution of alleles (using gda) showed that all loci were in HWE except WGA72 (FIS = 0.12) (Tab. I). The number of alleles per locus ranged from 8 to 38, with an overall estimated allelic diversity of 18.08. The relationship between cumulative number of alleles and sample size increased sharply as individuals were added beginning with the largest diameter class cohort and proceeding to the smaller size classes. This result indicated that there were high levels of neutral genetic diversity at Long Hollow that were not concentrated in a few highly heterozygous individuals, and that the alleles were shared across all size (Fig. 2) and value (Fig. 3) cohorts.
The average dbh for the Long Hollow black walnut population was 38.1 cm, with a range from less than 1.0 to 72.6 cm. The distribution of dbh for the total population produced a bell-shaped histogram, indicating an even-aged stand (Fig. 2). None of the four simulated diameter-limit harvests produced a reduction in overall allelic richness significantly different from that expected under a random harvest of the same intensity (Tab. II). While there was a 15% drop in the number of alleles under the most intense simulated treatment (> 30 cm cut), the change in Ar for that simulated harvest was not different from what would be expected from a random harvest of the same intensity. The estimated level of genetic differentiation (FST) between the pre- and post-harvest populations was close to zero and not significant, as was the genetic distance estimate (D) for all treatment levels (Tab. II). Both of these population parameter estimates indicate a high level of genetic similarity between the pre- and post-harvest populations.
Effect of simulated diameter limit harvests on genetic diversity and their level of significance. Before treatment level of allelic richness for the Long Hollow walnut population was 18.08 across all loci with a total 217 alleles.
Effect of simulated value-based harvest treatments on genetic diversity and their level of significance. Before treatment level of allelic richness for the Long Hollow walnut population was 18.08 across all loci with a total 217 alleles.
The average total dollar value of the 202 black walnut trees as evaluated by three foresters was $64867 and ranged from $62666 to $68066 (using log market values from 2003). None of the four simulated value-based (high grading) harvests produced a change in overall allelic richness different from that of a random harvest of the same intensity. The most severe simulated value-based treatment would have removed the most valuable 33% of the trees. This would have resulted in only a slight change in the allelic richness of the stand (Ar = 17.58 after treatment vs. Ar = 18.08 before) and would have produced a total loss of only 6 alleles (Tab. III). As in the simulated diameter-limit cuts, both differentiation statistics, FST and Nei’s D, indicated a high level of genetic similarity between the pre- and post-harvest populations (Tab. III).
We found no association between cohort dbh and the number of alleles private to the cohort. For example, there were 79 trees in the 0 to 30 cm dbh cohort which contained eight private alleles (alleles not found in any other cohort); the cohort of trees larger than 60 cm contained 23 trees and two private alleles. In every size cohort a private allele was encountered in 5 to 10% of the trees. The most valuable trees (top 33%, see Fig. 3) had a significantly smaller proportion of private alleles than the least valuable 67% of the trees (4.2% versus 16.3% respectively, two-sided P < 0.0035). The average observed heterozygosity for the size (dbh) cohorts was 0.796 ± 0.017, only the largest (> 60 cm) and smallest (< 30 cm) cohorts had an observed heterozygosity greater than the mean. Observed heterozygosity increased slightly but continuously with cohort value, from 0.786 (for the least valuable 67% of the trees) to 0.824 (for the most value 10% cohort).
4. Discussion
Analyses of the microsatellite data indicate that the walnuts in Long Hollow had high levels of genetic diversity (Ar = 18.08; expected heterozygosity, HE = 0.808). Results from studies of other black walnut populations have found high levels of genetic diversity in black walnut seeds using allozymes (HE = 0.22 − 0.34) (Busov et al., 2002; Rink et al., 1989; Rink et al., 1994) and adult trees using microsatellites (Robichaud et al., 2006; Victory et al., 2006). For example, Victory et al. (2006) observed comparable levels of genetic diversity (Ar = 22.91; HE = 0.793) averaged across loci in a range-wide survey of 43 populations of black walnut using the same 12 microsatellites used in this study. Robichaud et al. (2006) estimated similar levels of genetic diversity to those reported here (Ar = 19.90, HE = 0.832) in a 40 year-old black walnut provenance/progeny trial of 39 open-pollinated families representing a broad sampling from throughout the native range of black walnut. Estimates of Ar and HE based on microsatellite data for other temperate hardwood tree species were similar to those observed in the Long Hollow black walnut population (Dow and Ashley, 1996; van der Schoot et al., 2000).
In the present study, all loci were in HWE except WGA72 (FIS = 0.12). The fixation index estimate for WGA72 was moderate. Although a significant heterozygote deficiency can indicate assortative mating (inbreeding), low levels of pollen-gene flow, or null alleles, since only one locus was out of HWE, it is most likely that the observed deviation from HWE was caused by the presence of null alleles. Over all loci, we observed a small and non-significant heterozygote deficiency (FIS = 0.020) in the Long Hollow black walnut population, indicating that it is very close to HWE and therefore undergoing random mating.
None of the simulated harvests for either the value-based or diameter-limit cuts decreased levels of allelic richness significantly more than would be expected after a random harvest of equal intensity. In fact, for both types of simulated harvests investigated and across all treatments except one, the resulting reduction in allelic richness ranged from one to six percent. Only the diameter-limit simulated harvest removing all trees > 30 cm, a 70% reduction in population, substantially decreased the overall number of alleles, a loss of 15%. The decrease in allelic richness was not significantly different from a random harvest of equal intensity, but it can be argued that a reduction in allele numbers of 15% would be a biologically significant loss of diversity. To mitigate this loss of allelic richness, timing the harvest to coincide after a good nut crop would encourage natural regeneration and help insure a smaller genetic impact to the remnant population.
Overall, it appears that for this population, the allelic diversity was not over-concentrated in the largest or most valuable trees, but instead was evenly distributed across the entire population. We suggest several explanations of the observed results. First, roughly 80 years ago, when the Long Hollow site was first allowed to revert back to forest, it was a patchwork of open fields interspersed with small, early successional forest fragments. Black walnut, as a shade-intolerant species, needs large open areas for its seeds to germinate and for successful recruitment and establishment. The presence of open areas at Long Hollow during the time the forest was regenerating produced abundant opportunities for recruitment and establishment of the founder’s progeny. Second, no commercial harvesting or thinning has been conducted on the study site since it was acquired by the USDA Forest Service, allowing roughly 60 y of undisturbed growth and reproduction. So not only was there space for successful regeneration, but there also was plenty of time for the founders to reproduce. Parentage data from a concurrent study of the Long Hollow black walnut population support this hypothesis (unpublished data).
High levels of gene flow, specifically pollen-gene flow, played a role in the observed results. Gene flow from nearby but slightly differentiated populations can mitigate gene loss caused by small effective population size, natural and artificial selection, or genetic drift (Hamrick and Nason, 2000; Namkoong et al., 2000) by replacing lost genes or adding new ones to a population. There is a growing body of evidence in walnut (Busov et al., 2002; Polito et al., 2005; Robichaud et al., 2006; Victory et al., 2006) and in other wind-pollinated species (Bacles et al., 2005; Imbert and Lefevre, 2003; Valbuena-Carabana et al., 2005) that pollen gene dispersal occurs over hundreds to thousands of meters across varied landscapes. Therefore the relatively homogeneous distribution of genetic diversity across size cohorts observed in the Long Hollow population probably derived from moderate to high levels of pollen immigration. Victory et al. (2005) found that even the lowest frequency alleles in black walnut were not private to sub-populations but regionally distributed. Given the high rates of gene flow predicted for black walnut, we expect that alleles lost to harvest are often restored in the regenerating cohorts by pollen immigration, as was observed for Douglas-fir (Neale, 1985). Thus, long-term retention of genetic diversity in black walnut, a shade intolerant species, may depend on maintaining high levels of recruitment, as was observed by Degen et al. (2006) in a study of four tropical species.
We did not observe an increase in heterozygosity with tree size, as was reported by Nijensohn et al. (2005), nor did we observe any differences among the size cohorts for numbers of private alleles. The largest trees and smallest trees had about the same level of heterozygosity and proportion of private alleles. A slightly different picture emerged, however, when the Long Hollow population was divided into value cohorts. Although not directly investigated by our study, our data could support the observation by Nijensohn et al. (2005) that thinning based on value removes more rare alleles than random thinning. The genetic consequences of a loss of rare alleles from a single population can be hard to predict because rare alleles may be detrimental to adaptation in the short term or locally, but advantageous in the long-term or regionally.
It appears that for this particular stand of black-walnut, allelic richness was evenly distributed across the population. Removal of the largest and most valuable trees would not be expected to reduce the level of neutral genetic variation within the population to a greater degree than would a harvest of the same number of trees at random. It is likely that the ecology of black walnut and the stand history of Long Hollow interacted to produce the observed results. More than 60 y has passed since the last commercial harvests at Long Hollow, providing opportunities for the “founders” to successfully pass on their alleles to the younger cohorts. Finally, pollen-mediated gene flow most likely connected Long Hollow to other, distant populations, helping to maintain (or even increase) the allelic richness of the Long Hollow black walnut population over time. Ongoing investigation into realized and potential pollen-mediated gene flow for black walnut will provide more empirical support for this last hypothesis.
Although this study indicates that harvest scenarios are expected to have minimal impacts on the genetic diversity of black walnut in locations such as Long Hollow, the simulation in this study was for one cutting cycle only, and one population on one site. One could speculate that several sequential selective harvests of this site might eventually have strong demographic effects, but the negative genetic consequences of a small founder population on subsequent genetic diversity in a stand would probably depend strongly on rates of pollen gene flow. Previously published data support very low levels of genetic structure at even regional scales in black walnut (Victory et al., 2006). Because this study did not consider an actual harvest, we could not account for the stochastic processes and evolutionary forces that might affect allele frequency as natural regeneration takes place. These results should therefore be considered preliminary, and not readily generalizable. Studies of the intergenerational genetic consequences of harvest practices would be especially useful for understanding how to manage and maintain genetic diversity in forests over the long term.
Acknowledgments
We are indebted to B. Beheler, D. Carlson, and M. Cogswell, who graded the black walnut stand at Long Hollow. Without their volunteer efforts, much of this study would have been impossible. We also would like to thank the USDA Forest Service for access to the Long Hollow to conduct our study. Special thanks to D.Weigel for his in depth knowledge of the site. Funding for this project was provided by the USDA Forest Service, Purdue University, the Indiana Hardwood Lumbermen Association, the National Hardwood Lumbermen Association and the van Eck Foundation.
References
- Aldrich P.R. and Hamrick J.L., 1998. Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 281: 103–105. [CrossRef] [PubMed] [Google Scholar]
- Aldrich P.R.,Parker G.R.,Michler C.H., and Romero-Severson J., 2003. Whole-tree silvic identifications and the microsatellite genetic structure of a red oak species complex in an Indiana old-growth forest. Can. J. For. Res.-Rev. Can. Rech. For. 33: 2228–2237. [CrossRef] [Google Scholar]
- Aldrich P.R.,Parker G.R.,Romero-Severson J., and Michler C.H., 2005. Confirmation of oak recruitment failure in Indiana old-growth forest: 75 years of data. For. Sci. 51: 406–416. [Google Scholar]
- Angers V.A.,Messier C.,Beaudet M., and Leduc A., 2005. Comparing composition and structure in old-growth and harvested (selection and diameter-limit cuts) northern hardwood stands in Quebec. For. Ecol. Manage. 217: 275–293. [CrossRef] [Google Scholar]
- Bacles C.F.E.,Burczyk J.,Lowe A.J., and Ennos R.A., 2005. Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus excelsior L. Evolution 59: 979–990. [PubMed] [Google Scholar]
- Beineke W.F. 1974. Recent changes in the population structure of black walnut. In: R.B. Polk (ed.), Proceedings, Eighth Central States Forest Tree Improvement Conference, 43–46. University of Missouri, Columbia, Missouri, USA, [Google Scholar]
- Bekessy S.A.,Ennos R.A.,Burgman M.A.,Newton A.C., and Ades P.K., 2003. Neutral DNA markers fail to detect genetic divergence in an ecologically important trait. Biol. Conserv. 110: 267–275. [CrossRef] [Google Scholar]
- Boshier D.H., 2000. Mating systems. In: Young A., Boshier D., and Boyle T. (Eds.), Forest Conservation Genetics. CSIRO Publishing, Collingwood, pp. 63–79. [Google Scholar]
- Bratkovich S., Gallion J., Leatherberry E., Hoover W., Reading W., and Durham G., 2004. Forests of Indiana: their economic importance. In: US Department of Agriculture, Forest Service, Northeastern Area State and Private Forestry, St Paul, MN, pp. 1–18. [Google Scholar]
- Buchert G.P.,Rajora O.P.,Hood J.V., and Dancik B.P., 1997. Effects of harvesting on genetic diversity in old growth eastern white pine in Ontario, Canada. Conserv. Biol. 11: 747–758. [CrossRef] [Google Scholar]
- Busov V.B.,Rink G., and Woeste K., 2002. Allozyme variation and mating system of black walnut (Juglans nigra L.) in the Central Hardwood region of the United States. For. Genet. 9: 319–326. [Google Scholar]
- Clatterbuck W.K. 2006. Treatments for improving degraded hardwood stands. Professional Hardwood Notes #6. Knoxville, TN: University of Tennessee Extension SP680, 12 p. [Google Scholar]
- Collevatti R.G.,Grattapaglia D., and Hay J.D., 2001. Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Mol. Ecol. 10: 349–356. [CrossRef] [PubMed] [Google Scholar]
- Dow B.D. and Ashley M.V., 1996. Microsatellites analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol. Ecol. 5: 615–627. [CrossRef] [Google Scholar]
- Dow B.D. and Ashley M.V., 1998. Factors influencing male mating success in bur oak, Quercus macrocarpa. New For. 15: 161–180. [CrossRef] [Google Scholar]
- Finkeldey R. and Ziehe M., 2004. Genetic implications of silvicultural regimes. For. Ecol. Manage. 197: 231–244. [CrossRef] [Google Scholar]
- Glaubitz J.C., Robichaud R.L., Woeste K., Olin E., and Rhodes J., 2004. Toward guidelines for harvest intensities with minimal impact upon retained genetic diversity in Central Hardwood tree species. In: Proceedings of the 14th Central Hardwoods Forest Conference. US Department of Agricultural, Forest Service, Northeastern Research Station, Gen. Tech. Rep. NE-316, Wooster, OH March 16–19, pp. 395–403. [Google Scholar]
- Glaubitz J.C.,Wu H.X., and Moran G.F., 2003. Impacts of silviculture on genetic diversity in the native forest species Eucalyptus sieberi. Conserv. Genet. 4: 275–287. [CrossRef] [Google Scholar]
- Hamrick J.L. and Nason J.D., 2000. Gene flow in forest trees. In: Young A., Boshier D., Boyle T. (Eds.), Forest conservation genetics. CSIRO Publishing, Collingwood, pp. 81–90. [Google Scholar]
- Hawley G.J.,Schaberg P.G.,DeHayes D.H., and Brissette J.C., 2005. Silviculture alters the genetic structure of an eastern hemlock forest in Maine, USA. Can. J. For. Res. 35: 143–150. [CrossRef] [Google Scholar]
- Imbert E. and Lefevre F., 2003. Dispersal and gene flow of Populus nigra (Salicaceae) along a dynamic river system. J. Ecol. 91: 447–456. [CrossRef] [Google Scholar]
- Jones A.G. and Ardren W.R., 2003. Methods of parentage analysis in natural populations. Mol. Ecol. 12: 2511–2523. [CrossRef] [PubMed] [Google Scholar]
- Kelty M.J.,Kittredge D.B.,Kyker-Snowman T., and Leighton A.D., 2003. The conversion of even-aged stands to uneven-aged structure in southern New England. North. J. Appl. For. 20: 109–116. [Google Scholar]
- Kenefic L.S.,Sendak P.E., and Brissette J.C., 2005. Comparison of fixed diameter-limit and selection cutting in northern conifers. North. J. Appl. For. 22: 77–84. [Google Scholar]
- Lee C.-T.,Wickneswari R.,Mahani M.C., and Zakri A.H., 2002. Effect of selective logging on the genetic diversity of Scaphium macropodum. Biol. Conserv. 104: 107–118. [CrossRef] [Google Scholar]
- Lefort F. and Douglas G.C., 1999. An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Ann. For. Sci. 56: 259–263. [CrossRef] [EDP Sciences] [Google Scholar]
- Lewis P.O. and Zaykins D., 2001. Genetic data analysis: Computer program for the analysis of allelic data. In: p. Free program distributed by the authors over the internet from http:lewis.eeb.uconn.edu/lewishome/software.html. [Google Scholar]
- Marquardt P.E. and Epperson B.K., 2004. Spatial and population genetic structure of microsatellites in white pine. Mol. Ecol. 13: 3305–3315. [CrossRef] [PubMed] [Google Scholar]
- McGuire M.,Stevens J., and Potter-Witter K., 1999. Assessing scarcity of the north central veneer log resource. North. J. Appl. For. 16: 160–166. [Google Scholar]
- Murawski D.A.,Gunatilleke I., and Bawa K.S., 1994. The effects of selective logging on inbreeding in Shorea megistophylla (Dipterocarpaceae) from Sri-Lanka. Conserv. Biol. 8: 997–1002. [CrossRef] [Google Scholar]
- Namkoong G., Koshy M.P., and Aiken S., 2000. Selection. In: Young A., Boshier D., and Boyle T. (Eds.), Forest conservation genetics. CSIRO Publishing, Collingwood, pp. 101–111. [Google Scholar]
- Neale D.B., 1985. Genetic implications of shelterwood regeneration of Douglas-fir in southwest Oregon. For. Sci. 31: 995–1005. [Google Scholar]
- Nei M., 1973. Analysis of Gene Diversity in Subdivided Populations. Proc. Nat. Acad. Sci. 70: 3321–3323. [Google Scholar]
- Nijensohn S.E.,Schaberg P.G.,Hawley G.J., and DeHayes D.H., 2005. Genetic subpopulation structuring and its implications in a mature eastern white pine stand. Can. J. For. Res. 35: 1041–1052. [CrossRef] [Google Scholar]
- Nyland R.D., 2005. Diameter-limit cutting and silviculture: A comparison of long-term yields and values for uneven-aged sugar maple stands. North. J. Appl. For. 22: 111–116. [Google Scholar]
- Obayashi K.,Tsumura Y.,Ihara-Ujino T.,Niiyama K.,Tanouchi H.,Suyama Y.,Washitani I.,Lee C.T.,Lee S.L., and Muhammad N., 2002. Genetic diversity and outcrossing rate between undisturbed and selectively logged forests of Shorea curtisii (Dipterocarpaceae) using microsatellite DNA analysis. Int. J. Plant Sci. 163: 151–158. [CrossRef] [Google Scholar]
- Parker P.G.,Snow A.A.,Schug M.D.,Booton G.C., and Fuerst P.A., 1998. What molecules can tell us about populations: Choosing and using a molecular marker. Ecology 79: 361–382. [Google Scholar]
- Polito V.S.,Pinney K.,Weinbaum S.,Aradhya M.K.,Dangl J.,Yanknin Y., and Grant J.A., 2005. Walnut pollination dynamics: pollen flow in walnut orchards. Acta Hortic. 705: 465–472. [Google Scholar]
- Rajora O.P., 1999. Genetic biodiversity impacts of silvicultural practices and phenotypic selection in white spruce. Theor. Appl. Genet. 99, 954–961. [CrossRef] [Google Scholar]
- Rajora O.P. and Rahman M.H., 2003. Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus × canadensis) cultivars. Theor. Appl. Genet. 106: 470–477. [PubMed] [Google Scholar]
- Rajora O.P.,Rahman M.H.,Buchert G.P., and Dancik B.P., 2000. Microsatellite DNA analysis of genetic effects of harvesting in old-growth eastern white pine (Pinus strobus) in Ontario, Canada. Mol. Ecol. 9: 339–348. [CrossRef] [Google Scholar]
- Rink G.,Carroll E.R., and Kung F.H., 1989. Estimation of Juglans nigra L. mating system parameters. For. Sci. 35: 623–627. [Google Scholar]
- Rink G.,Zhang G.,Jinghua Z.,Kung F.H., and Carroll E.R., 1994. Mating parameters in Juglans nigra L. seed orchard similar to natural population estimates. Silvae Genet. 43: 261–263. [Google Scholar]
- Robichaud R.L.,Glaubitz J.C.,Olin E.,Rhodes J., and Woeste K., 2006. A robust set of black walnut microsatellites for parentage and clonal identification. New For. 32: 179–196. [CrossRef] [Google Scholar]
- Sork V.L.,Smouse P.E.,Apsit V.J.,Dyer R.J., and Westfall R.D., 2005. A two-generation analysis of pollen pool genetic structure in flowering dogwood, Cornus florida (Cornaceae), in the Missouri Ozarks. Am. J. Bot. 92: 262–271. [CrossRef] [PubMed] [Google Scholar]
- Streiff R.,Ducousso A.,Lexer C.,Steinkellner H.,Gloessl J., and Kremer A., 1999. Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q. petraea (Matt.) Liebl. Mol. Ecol. 8: 831–841. [CrossRef] [Google Scholar]
- Tormoehlen B., Gallion J., and Schmidt T.L., 2000. Forests of Indiana: a 1998 overview. In: US Department of Agriculture, Forest Service, Northeastern Area State and Private Forestry, St Paul, MN, pp. 1–24. [Google Scholar]
- Tritton L.M. and Sendak P.E., 1995. Ecological aspects of forest management planning: a Northern hardwood forest case study. North. J. Appl. For. 12: 121–126. [Google Scholar]
- Valbuena-Carabana M.,Gonzalez-Martinez S.C.,Sork V.L.,Collada C.,Soto A.,Goicoechea P.G., and Gil L., 2005. Gene flow and hybridisation in a mixed oak forest (Quercus pyrenaica Willd. and Quercus petraea (Matts.) Liebl.) in central Spain. Heredity 95: 457–465. [CrossRef] [PubMed] [Google Scholar]
- Van der Schoot J.,Pospiskova M.,Vosman B., and Smulders M.J.M., 2000. Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Theor. Appl. Genet. 101: 317–322. [CrossRef] [Google Scholar]
- Victory E., Woeste K., and Rhodes O.E. Jr., 2004. History of black walnut genetics research in North America. In: Michler C.H. et al. (Eds.), Black walnut in a new century, proceedings of the 6th Walnut Council research symposium, 2004 July 25–28, Lafayette, IN. Gen. Tech. Rep. NC-243. St. Paul, MN: US Department of Agriculture, Forest Service, North Central Forest Experiment Station: 1–8. [Google Scholar]
- Victory E.,Glaubitz J.C.,Olin E.,Rhodes J., and Woeste K.E., 2006. Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am. J. Bot. 93, 118–126. [CrossRef] [Google Scholar]
- Weir B.S. and Cockerham C.C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [CrossRef] [PubMed] [Google Scholar]
- Ziehe M. and Hattemer H.H., 2002. Target-Diameter felling and consequences for genetic structures in a beech stand (Fagus sylvatica L.). In: von Gadow K., Nagel J., and Saborowski J. (Eds.), Continuous cover forestry, assessment, analysis, scenarios. Kluwer Academic Publishers, Dordrecht, Netherlands, pp. 91–105. [Google Scholar]
All Tables
Properties of the 12 microsatellite markers employed in this study. Genbank accession* numbers are in parentheses under the locus name. The number in parentheses under the allele size range is the number of alleles found for that locus in the Long Hollow black walnut population. The FIS values for the black walnut population is based on a total census of the Long Hollow stand.
Effect of simulated diameter limit harvests on genetic diversity and their level of significance. Before treatment level of allelic richness for the Long Hollow walnut population was 18.08 across all loci with a total 217 alleles.
Effect of simulated value-based harvest treatments on genetic diversity and their level of significance. Before treatment level of allelic richness for the Long Hollow walnut population was 18.08 across all loci with a total 217 alleles.
All Figures
 |
Figure 1 (A) Map showing the Long Hollow study area in the Hoosier National Forest HNF, southern Indiana, USA, (inset shows larger view of the area). (B) Location of all 278 black walnut trees sampled at the Long Hollow research site. |
| In the text | |
 |
Figure 2 Diameter breast height class distribution of 278 black walnut trees in Long Hollow. Four simulated diameter-limit harvests (30, 40, 50, 60 cm) were performed, removing all individuals to the right of the arrows. Right axis indicates cumulative number of alleles in the population. |
| In the text | |
 |
Figure 3 Distribution of the estimated value of the Long Hollow black walnuts. Four simulated value-based harvests were performed by removing the most-valuable 10, 15, 25, and 33% of all the trees from the population. Right axis indicates cumulative number of alleles found in the Long Hollow population. |
| In the text | |
 |
Figure 4 An example null distribution of allelic richness (Ar) for the simulated diameter-limit harvest of > 50 cm. The distribution was generated by performing 10000 random harvests of equal intensity each with an equal number of “harvested” trees chosen at random with respect to diameter. After each simulated harvest, allelic diversity was recalculated based on the genotypes of the residual black walnuts, and the observed value for the > 50 cm harvest was then compared to this null distribution to obtain its statistical significance (one-tail test). All other simulated diameter-limit and value-based harvests generated similarly-shaped distributions and comparable P values, so only this example distribution is shown. The shadowed part of the histogram represents the amount of allelic richness (Ar) of the residual trees after the > 50 cm harvest was conducted. |
| In the text | |


